
Written by Jean-François Mangin, Clément Langlet, Antoine Grigis, Vincent Frouin and Denis Rivière.
The structural connectome is one of the cornerstones of research programs simulating the dynamics of brain function. This connectome is a matrix that represents the connections between the different brain areas. For the past twenty years, it has been possible to access a macroscopic version of this connectome from diffusion MRI, which allows scientists to map the brain fiber bundles in vivo (cf Fig. 1). It is now common to map millions of bundle trajectories for each brain. Hence, studies that require the comparison of thousands of brains require significant computational resources.
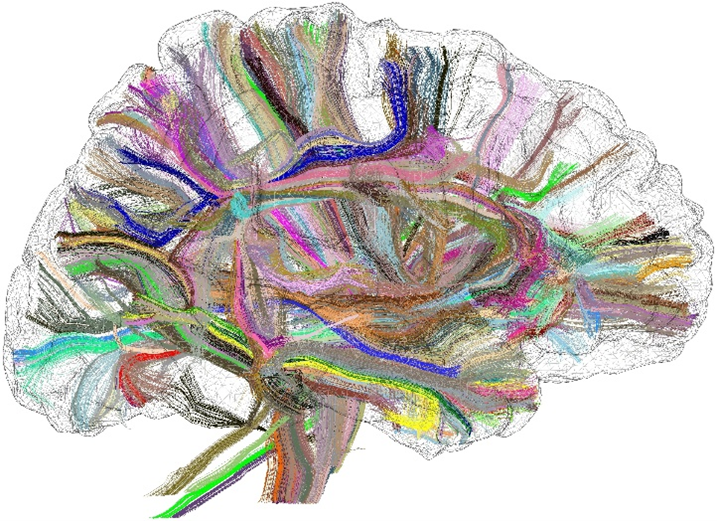
Figure 1 : Long fiber bundles mapped with diffusion MRI.
The macroscopic structural connectome of a brain is obtained for a parcellation of the gray matter, which includes the cell bodies of the neurons. The matrix is filled up by the connections between these parcels established by the bundles organizing the neuron axons. A wide variety of parcellations can be used, depending on the questions investigated. The chosen parcellation is usually projected onto each brain in a study using a registration operation based on the brain morphology, especially cortical folding. But since there is no absolute link between morphology and fiber organization, this approach is suboptimal.
More than 1000 individuals fibers organitzation analyzed
Thanks to the resources of Fenix, a team from Neurospin, CEA, has analyzed the organization of the fibers of 1000 individuals of the American Human Connectome Project database, which provides outstanding diffusion MRI data (www.humanconnectome.org). This analysis, based on the Constellation software (Lefranc et al., brainvisa.info), has yielded an optimal parcellation of the cortex, in the sense that each parcel corresponds to a region whose connectivity to the rest of the brain is stable throughout the population (see Fig. 2). Machine learning tools were used to adapt this average parcellation to the organization of the fibers of each individual, to obtain very stable connectomes from one individual to another (see Fig. 3). These optimal individual parcellations were then used to create a functional connectome that describes probable communications between these parcels from functional MRI images. For each of the 1000 individuals, the structural and functional connectomes have been made available to the modelling community (Langlet et al.). They will also give rise to heritability studies.

Figure 2 Optimal parcellation of the cortex of 1000 individuals based on structural connectivity
Figure 3 : Machine-learning-based tuning of the parcellation to individuals to achieve stable structural connectomes
- Lefranc S. et al., Groupwise connectivity-based parcellation of the whole human cortical surface using watershed-driven dimension reduction. Med Image Anal. 2016 30:11-29. doi: 10.1016/j.media.2016.01.003.
- Langlet, C., Rivière, D., & Mangin, J.-F. (2022). Nested parcellations connectome delivered for one large dataset using Constellation algorithm (v1.1) [Data set]. EBRAINS. https://doi.org/10.25493/5RCR-GS
Acknowledgements
We acknowledge the use of Fenix Infrastructure resources, which are partially funded from the European Union’s Horizon 2020 research and innovation programme through the ICEI project under the grant agreement No. 800858. This study was further supported by the European Union’s Horizon 2020 Framework Programme for Research and Innovation under Specific Grant Agreements No. 945539 (HBP SGA3) and No. 785907 (HBP SGA2), the Priority Program 2041 (SPP 2041) “Computational Connectomics” (DFG), the Helmholtz IVF Grant SO-092 (ACA), the Joint Lab SMHB, and the INFN APE Parallel/Distributed Computing laboratory.
HCP Data were provided by the Human Connectome Project, WU-Minn Consortium (Principal Investigators: David Van Essen and Kamil Ugurbil; 1U54MH091657) funded by the 16 NIH Institutes and Centers that support the NIH Blueprint for Neuroscience Research; and by the McDonnell Center for Systems Neuroscience at Washington University.