
Written by Gianmarco Tiddia, Bruno Golosio, Jose Villamar, Johanna Senk, Jasper Albers, Francesco Simula, Jari Pronold, Viviana Fanti, Elena Pastorelli, Pier Stanislao Paolucci, Sacha Jennifer van Albada
Spiking neural network models have become an effective tool to study brain functions as they capture several aspects of natural neural networks. Every neuron in such a network model is characterized by a membrane potential and a mechanism to emit electrical pulses for the communication with other neurons. The multi-area model of macaque visual cortex is a large-scale spiking network model describing the dynamics of 32 vision-related areas of the macaque cortex at cellular resolution [1-3]. Each area has a laminar structure (see Figure 1) and in total the model is composed of 4.1 million neurons and about 24 billion synapses. Integrating experimental data on the network architecture of all represented areas, the model is able to reproduce the resting-state activity experimentally observed in the macaque brain.
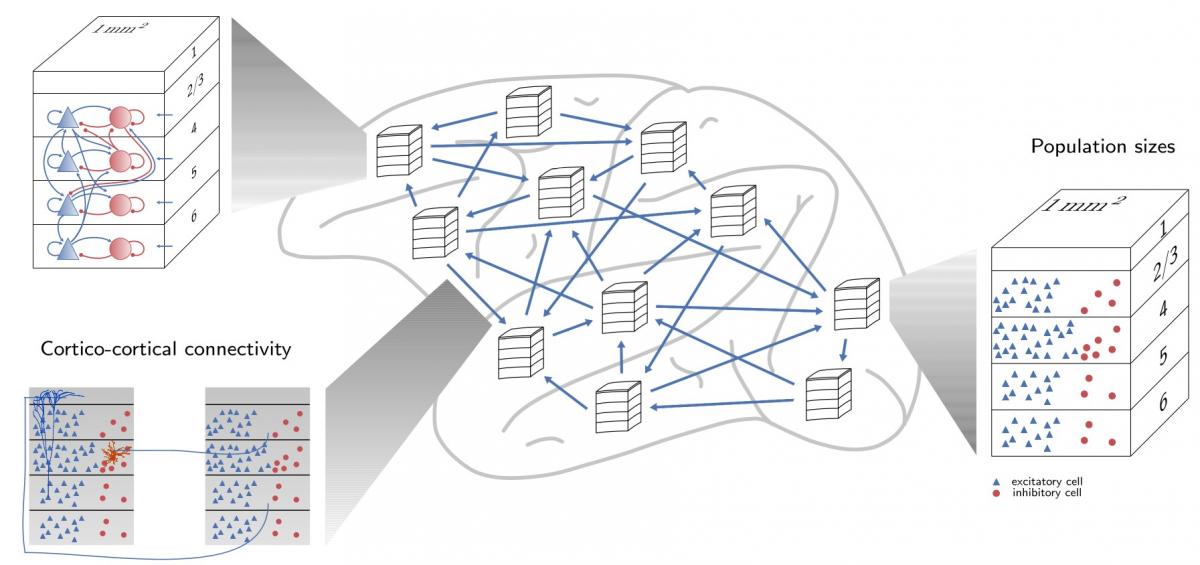
Figure 1: A schematic overview of the multi-area model. Each area has a laminar structure, with one excitatory and one inhibitory neuron population in each layer.
Simulating such large-scale models is computationally demanding. Indeed, achieving reasonable simulation times is hard even taking advantage of high-performance computing (HPC) clusters. In addition, performant software tools are needed to efficiently exploit the computational resources. Simulators such as NEST [4] can simulate a plethora of neuron and synapse models, enabling large-scale simulations on CPU-based systems. Further, the interest in GPU-based simulations of spiking neural networks is rising since the growth of the GPU industry leads to more and more performant and accessible GPU cards with an increased amount of memory. Therefore there is a need for simulators capable of exploiting this fast-growing hardware.
NEST GPU [5-7] is a library recently integrated into the NEST Initiative to enable spiking simulations on GPUs in a framework similar to that of the NEST simulator.
Recent progress in the development of NEST GPU facilitates the simulation of large-scale network models distributed across multiple nodes of a compute cluster. We simulated the multi-area model with both NEST and NEST GPU to compare the results [7]. The simulations were performed on the HPC cluster JUSUF of the Jülich Supercomputing Center (JSC) made available via ICEI allocations for the Human Brain Project (HBP). We observed that the two simulators show consistent results in terms of statistics of the neural activity, while the GPU-based simulator outperformed the CPU-based one by a factor 3.1 in terms of simulation time, simulating one second of biological time of the model in less than 16 s of wall-clock time.
This work opens up the possibility of exploiting GPU clusters to perform fast and reliable simulations of very large-scale spiking networks of brain models.
References
[1] Schmidt M, Bakker R, Hilgetag CC, Diesmann M & van Albada SJ (2018), Multi-scale account of the network structure of macaque visual cortex, Brain Structure and Function, https://doi.org/10.1007/s00429-017-1554-4
[2] Schuecker J, Schmidt M, van Albada SJ, Diesmann M & Helias M (2017), Fundamental Activity Constraints Lead to Specific Interpretations of the Connectome, PLOS Computational Biology , https://doi.org/10.1371/journal.pcbi.1005179
[3] Schmidt M, Bakker R, Shen K, Bezgin B, Diesmann M & van Albada SJ (2018), A multi-scale layer-resolved spiking network model of resting-state dynamics in macaque cortex, PLOS Computational Biology, https://doi.org/10.1371/journal.pcbi.1006359
[4] Gewaltig MO, Diesmann M: NEST (NEural Simulation Tool). Scholarpedia 2007, 2(4):1430, http:/doi.org/10.4249/scholarpedia.1430
[5] Source code: https://github.com/nest/nest-gpu
[6] Golosio B, Tiddia G, De Luca C, Pastorelli E, Simula F & Paolucci PS (2021), Fast simulations of highly-connected spiking cortical models using GPUs, Frontiers in Computational Neuroscience, https://doi.org/10.3389/fncom.2021.627620
[7] Tiddia G, Golosio B, Albers J, Senk J, Simula F, Pronold J, Fanti V, Pastorelli E, Paolucci PS & van Albada SJ (2022), Fast simulation of multi-area spiking network model of macaque cortex on an MPI-GPU cluster, Frontiers in Neuroinformatics, https://doi.org/10.3389/fninf.2022.883333
Acknowledgements
We acknowledge the use of Fenix Infrastructure resources, which are partially funded from the European Union’s Horizon 2020 research and innovation programme through the ICEI project under the grant agreement No. 800858. This study was further supported by the European Union’s Horizon 2020 Framework Programme for Research and Innovation under Specific Grant Agreements No. 945539 (HBP SGA3) and No. 785907 (HBP SGA2), the Priority Program 2041 (SPP 2041) “Computational Connectomics” (DFG), the Helmholtz IVF Grant SO-092 (ACA), the Joint Lab SMHB, and the INFN APE Parallel/Distributed Computing laboratory.