
Written by Masoli, S., Rizza, M.F., De Schepper, R., Geminiani, A., Casellato, C., D’Angelo, E.
Department of Brain and Behavioural Sciences (University of Pavia), Human Brain Project, WP1
The ICEI projects ich003 and ich007 were granted for modelling and simulation of cerebellar neuron and network models (D’Angelo et al., 2016). The cerebellum is part of the central nervous system and endlessly processes a huge amount of information contributing to motor coordination, balance, language and high cognitive functions.
The cerebellar neuron models were built using the open-source NEURON simulation environment (Hines et al., 2009), the Python script language and the “Blue Brain Python Optimisation Library” (BluePyOpt) framework (Van Geit et al., 2016). In the Human Brain Project (HBP), NEURON allowed to model and simulate detailed multicompartmental neuron models of granule, Golgi, stellate and Purkinje cells. Importantly, a scaffold cerebellar network with realistic connectome was reconstructed using a new customized modelling framework, “The Brain Scaffold Builder” (BSB), hosting the multicompartmental neuron models to reproduce microcircuit processing and spatiotemporal dynamics.
The models were developed and experimentally constrained in the laboratories of the Neurophysiology Unit, Department of Brain and Behavioural Sciences (University of Pavia) under the supervision of Professor Egidio D’Angelo, leader of the HBP subproject on Rodent Brain Models “RisingNet”. The Fenix resources, granted through the PRACE-ICEI Call, allowed the modelling and simulation of the intrinsic and synaptic responses of cerebellar neurons inside their microcircuit, uncovering behaviours that could not be directly studied using experimental procedures only.
The most numerous and smallest neurons of the brain are the granule cells populating the cerebellar cortex with over 60% of all the brain neurons. Since many decades, the granule cells were thought to form a homogeneous family only capable of stereotypical responses. Surprisingly, models generated three types of responses, which differed for the degrees of spike-frequency adaptation (absent, moderate, strong). These results warranted ad hoc experiments in mice, which not only confirmed the three discharge modalities anticipated by simulations but also showed an additional one, firing acceleration. This fourth modality was shown to involve TRPM4 channels. Indeed, with the insertion of TRPM4 channels coupled to intracellular calcium dynamics, the model nicely reproduced firing acceleration (Masoli et al., 2020).
The Golgi cells are large neurons, whose main role is to inhibit granule cells. The Golgi cell models were based on 3D morphological reconstructions and were endowed with the latest information about ionic channel gating kinetics and subcellular distribution. The models supported the importance of specific ionic channels on the dendrites and allowed to study a broad range of synaptic input patterns. The simulations unveiled the existence of dendritic coincidence detection mechanisms and spike-timing-dependent plasticity (Masoli et al., 2021).
The stellate cells reside in the upper part of the cerebellar cortex and inhibit the dendrites of Purkinje cells, the sole output of the cerebellar cortical network. Our simulations predicted that few synaptic inputs were enough to activate spike bursts that can, in turn, suppress Purkinje cell firing in a frequency-dependent manner implementing a low-pass filtering circuit (Rizza et al., 2021; Masoli and D’Angelo, 2017).
All these models, with their specific properties, were integrated into a large-scale network using the BSB framework. A new ground-truth about circuit organization was generated and, for the first time, the activation of cerebellar cortical columns emerged (De Schepper et al., 2021) (Fig. 1).
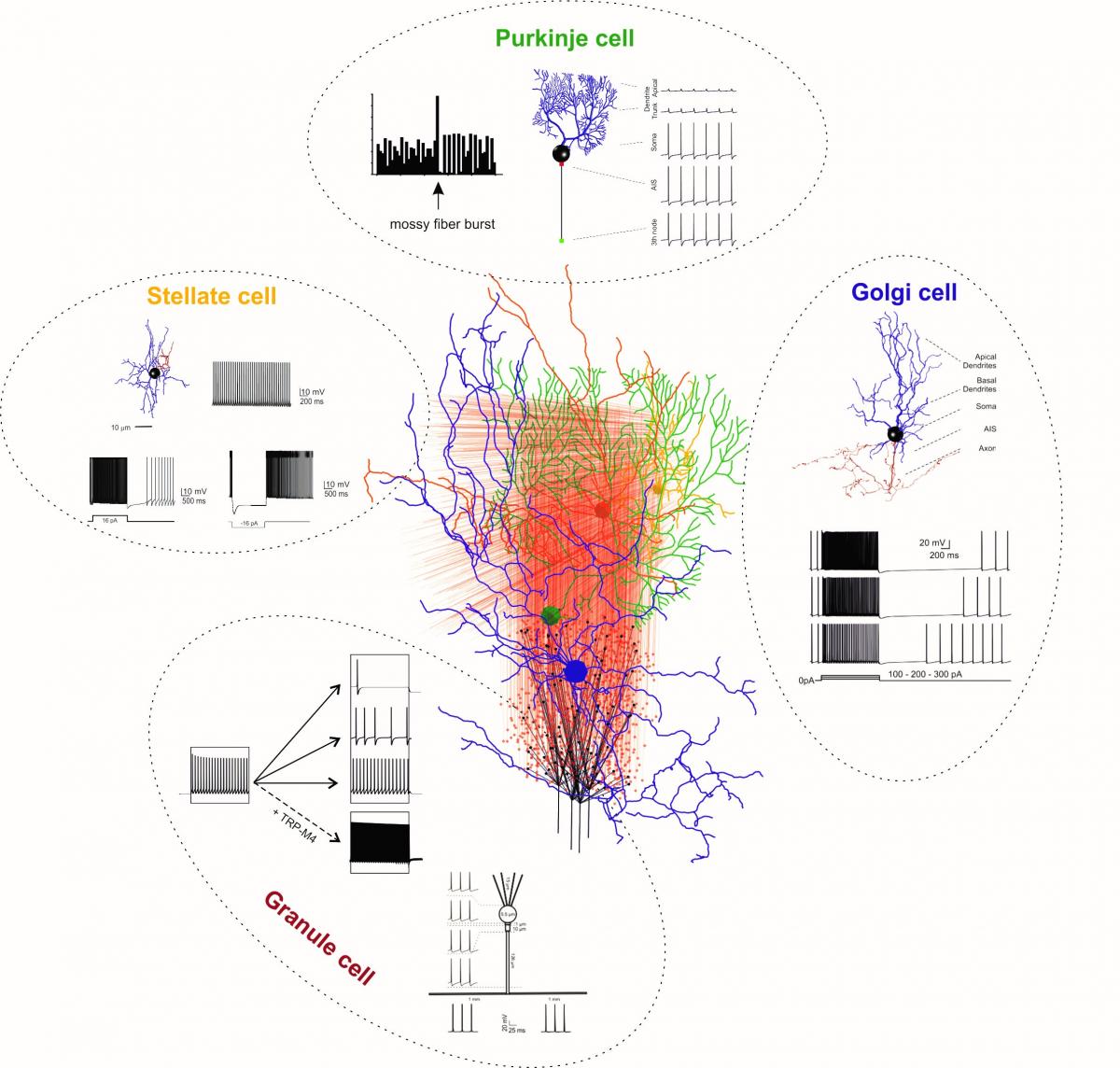 Figure 1: Detailed neuron models inserted into the cerebellar network. The cerebellar network is reconstructed using the Brain Scaffold Builder (De Schepper et al., 2021). Single neurons (granule, Golgi, stellate and Purkinje cells) are reconstructed using NEURON (the label names are in the same colour as neurons in the drawing). For each neuron type, simulations show the fundamental discharge properties during injection of depolarizing currents (details in Masoli et al., 2020; Rizza et al., 2021; Masoli et al., 2021, Masoli and D’Angelo, 2017). Simulations were run on PizDaint (Fenix).
Figure 1: Detailed neuron models inserted into the cerebellar network. The cerebellar network is reconstructed using the Brain Scaffold Builder (De Schepper et al., 2021). Single neurons (granule, Golgi, stellate and Purkinje cells) are reconstructed using NEURON (the label names are in the same colour as neurons in the drawing). For each neuron type, simulations show the fundamental discharge properties during injection of depolarizing currents (details in Masoli et al., 2020; Rizza et al., 2021; Masoli et al., 2021, Masoli and D’Angelo, 2017). Simulations were run on PizDaint (Fenix).
The Fenix infrastructure was used to support simulation workflows for the optimization and validation of multicompartmental single cell models and to reconstruct and simulate large-scale cerebellar networks. The computational requirements changed based on the size and complexity of neuron morphology, the types of ionic channels and synapses, the extension of the network and the stimulation protocols. The neuron reconstruction process made use of a genetic algorithm approach (BluePyOpt Framework) that required extensive calculation. With the computational resources provided by the Fenix Research Infrastructure via our ICEI projects, the BluePyOpt and the BSB frameworks allowed the simulation and validation of thousand models of neurons and networks on the HPC cluster PizDaint:
- Single neurons. Each optimization job required between 4 and 32 nodes (1152 cores) for a runtime of 1-5 hours.
- Networks. A simulation with ~30˙000 neurons and ~ 1˙500˙000 synapses lasting 8 secs of real-time network activity required 20 nodes (720 cores) for a runtime of ~6 hours.
In conclusion, FENIX-ICEI empowered the successful reconstruction and simulation of the cerebellar neurons and networks, which will now continue inside the “Cerebellar Modelling Hub” of the University of Pavia under the ESFRI-EBRAINS coordination (https://www.humanbrainproject.eu/en/collaborate/facility-hubs/).
References
1. D’Angelo E, Antonietti A, Casali S, Casellato C, Garrido JA, Luque NR, Mapelli L, Masoli S, Pedrocchi A, Prestori F, Rizza MF, Ros E. (2016). Modeling the Cerebellar Microcircuit: New Strategies for a Long-Standing Issue. Front Cell Neurosci.10:176. doi: 10.3389/fncel.2016.00176.
2. De Schepper, R., Geminiani, A., Masoli, S., Rizza, M.F., Antonietti, A., Casellato, C., D’Angelo, E. (2021). Scaffold modelling captures the structure-function-dynamics relationship in brain microcircuits. BiorXiv. doi: 10.1101/2021.07.30.454314
3. Hines, M. L., Davison, A. P., and Muller, E. (2009). NEURON and Python. Front. Neuroinform. 3, 1. doi: 10.3389/neuro.11.001.2009.
4. Masoli S, D’Angelo E. Synaptic (2017). Activation of a Detailed Purkinje Cell Model Predicts Voltage-Dependent Control of Burst-Pause Responses in Active Dendrites. Front. Cell. Neurosci., 13 September 2017 | https://doi.org/10.3389/fncel.2017.00278. PubMed PMID: 28303385
5. Masoli, S., Tognolina, M., Laforenza, U., Moccia, F., and D’Angelo, E. (2020). Parameter tuning differentiates granule cell subtypes enriching transmission properties at the cerebellum input stage. Commun. Biol. 3, 222. doi:10.1038/s42003-020-0953-x.
6. Masoli, S., Ottaviani, A., Casali, S., and D’Angelo, E. (2020). Cerebellar Golgi cell models predict dendritic processing and mechanisms of synaptic plasticity. PLoS Comput. Biol. 16, 1–27.doi:10.1371/journal.pcbi.1007937.
7. Rizza, M. F., Locatelli, F., Masoli, S., Sánchez-Ponce, D., Muñoz, A., Prestori, F., D’Angelo E., (2021). Stellate cell computational modeling predicts signal filtering in the molecular layer circuit of cerebellum. Sci. Rep. 11, 3873. doi:10.1038/s41598-021-83209-w.
8. Van Geit, W., Gevaert, M., Chindemi, G., Rössert, C., Courcol, J.-D., Muller, E. B., et al. (2016). BluePyOpt: Leveraging Open Source Software and Cloud Infrastructure to Optimise Model Parameters in Neuroscience. Front. Neuroinform. 10, 1–30. doi:10.3389/fninf.2016.00017.